CSE/EE 577: Medical Image AnalysisAutumn 2017 |
Download the images and data you need: hw2.zip.
 |
 |
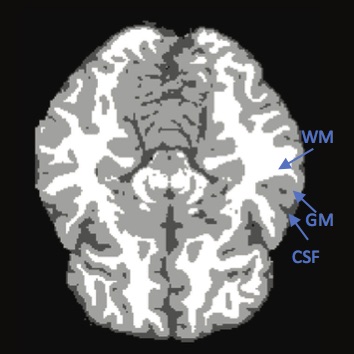
| a) sample brain MRI image | b) segmented image |
Brain MRI analysis is an essential task in many clinical applications. Fundamental components of structural brain MRI analysis include the classification of MRI data into specifict tissue types and the identification and description of specific anotomical structures. Among all the tissue types, the tissue elements we are typically interested in are: cerebrospinal fluid (CSF), gray matter (GM) and white matter (WM); see Figure b). The segmentation results are further used in different applications such as for analyzing anatomical structures, for studying pathological regions, for surgical planning, and for visualization purposes.
Brain image segmentation can be performed on 2D images, sequences of 2D images, or 3D volumetric images. There are several techniques to achieve the segmentation. They can be classfied into thresholding methods [1, 2, 3], clustering methods [4, 5, 6], edge detection methods [7, 8, 9], region-based methods[10, 11], model-based methods[12, 13, 14] and Hidden Markov Random Field methods [15, 16, 17].
In this assignment, we will try some basic 2D thresholding and clustering method to find the 3 interesting regions in the testing brain MRI images given 3 training images with their groundtruth labels. The MRI images and their corresponding labels are from the normal anatomical models simulated from BrainWeb (T1-weighted).
In addition to the training and testing images, we will also provide you with the other pre-labeled data. Feel free to use them as groundtruth to train other segmentation models.
Read in the 3 given training images and 3 corresponding labels. Cerebrospinal Fluid (CSF), label =1; Grey Matter (GM), label =2; White Matter (WM), label =3.
| clc;clear all;close all; %read in all the training images for i =1:3 trainImages(:,:,i)=imread(['train/',num2str(i),'.jpg']); load(['train/',num2str(i),'.mat']); trainLabels(:,:,i)=label; end [r,w,n]=size(trainImages); |
Here we calculate the mean CSF/GM/WM values for reference later.
| %% %calculate the mean CSF/GM/WM values meanCSF = mean(trainImages(trainLabels==1)); meanGM = mean(trainImages(trainLabels==2)); meanWM = mean(trainImages(trainLabels==3)); |
By observing the training and testing images, you might notice that besides the inner brain region, there are some outer non-brain regions such as skulls. It's hard to differentiate them from the inner brain region using only pixel intensities. Try to first remove the outer region using image morphology.
| %% % TODO hint: remove the outer brain region, consider using imerode/imdilate to %disconnect the inner and outer region. The inner brain region has more %pixels. Hint: consider using bwconncomp to extract the largest connecting region. testimage = imread('test/1.jpg'); mask = double(testimage> ); |
Cluster the remaining pixels using k-means method. You might need to explore the best k value.
| % TODO hint: use kmeans to cluster the pixels according to their
intensities [cluster_idx, cluster_center] |
cluster_center will give you the mean value of each cluster. Use the meanCSF/meanGM/meanWM values computed from training images to find the corresponding labels on the testing image. Assign label 1 to CSF, 2 to GM and 3 to WM. Visualize your segmentation result and compare it to the groundtruth provided.
| %% result = uint8(zeros(r,w)); % TODO hint: use the meanCSF/meanGM/meanWM value to find the corresponding %labels. load('test/1.mat'); |
Extra Credits
You are welcome to try other fancy techniques to segment and label the 3 parts. You can use the all the simulated brain images (data.zip) we provided as training data.
Homework is due on October 18 (Wednesday) by 11:59 PM PST.
This assignment can be done alone or in groups. If you do not know and/or do not have Matlab, we will pair you with someone who does, and you can get help from Shu.
References
[1] Kumar, S., et al.: Skull stripping and automatic segmentation of brain MRI using seed growth and threshold techniques pp. 422-426 (2007)
[2] Natarajan, P., Krishnan, N., Kenkre, N.S., Nancy, S., Singh, B.P.: Tumor detection using threshold operation in MRI brain images pp. 1-4 (2012)
[3] Zhao, M., Lin, H.Y., Yang, C.H., Hsu, C.Y., Pan, J.S., Lin, M.J.: Automatic threshold level set model applied on MRI image segmentation of brain tissue. Appl. Math 9(4), 1971-1980 (2015)
[4] Pham, D.L., Xu, C., Prince, J.L.: Current methods in medical image segmentation 1. Annual review of biomedical engineering 2(1), 315-337 (2000)
[5] Wu, Z., Leahy, R.: An optimal graph theoretic approach to data clustering: Theory and its application to image segmentation. Pattern Analysis and Machine Intelligence, IEEE Transactions on 15(11), 1101-1113 (1993)
[6] Chuang, K.S., Tzeng, H.L., Chen, S., Wu, J., Chen, T.J.: Fuzzy c-means clustering with spatial information for image segmentation. computerized medical imaging and graphics 30(1), 9-15 (2006)
[7] Perona, P., Malik, J.: Scale-space and edge detection using anisotropic diffusion. Pattern Analysis and Machine Intelligence, IEEE Transactions on 12(7), 629-639 (1990)
[8] Senthilkumaran, N., Rajesh, R.: Edge detection techniques for image segmentation-a survey of soft computing approaches. International journal of recent trends in engineering 1(2) (2009)
[9] Canny, J.: A computational approach to edge detection. Pattern Analysis and Machine Intelligence, IEEE Transactions on (6), 679-698 (1986)
[10] Lin, G.C., Wang, W.J., Kang, C.C., Wang, C.M.: Multispectral MR images segmentation based on fuzzy knowledge and modified seeded region growing. Magnetic resonance imaging 30(2), 230-246 (2012)
[11] Roura, E., Oliver, A., Cabezas, M., Vilanova, J.C., Rovira, A., Rami o-Torrent a, L., Llad o, X.: Marga: multispectral adaptive region growing algorithm for brain extraction on axial MRI. Computer methods and programs in biomedicine 113(2), 655-673 (2014)
[12] Chan, T.F., Vese, L., et al.: Active contours without edges. Image processing, IEEE transactions on 10(2), 266-277 (2001)
[13] Ho, S., Bullitt, L., Gerig, G.: Level-set evolution with region competition: automatic 3-d segmentation of brain tumors 1, 532-535 (2002)
[14] McInerney, T., Terzopoulos, D.: Deformable models in medical image analysis: a survey. Medical image analysis 1(2), 91-108 (1996)
[15] Held, K., Kops, E.R., Krause, B.J., Wells III, W.M., Kikinis, R., Muller-Gartner, H.W.: Markov random field segmentation of brain MR images. Medical Imaging, IEEE Transactions on 16(6), 878-886 (1997)
[16] Panjwani, D.K., Healey, G.: Markov random field models for unsupervised segmentation of textured color images. Pattern Analysis and Machine Intelligence, IEEE Transactions on 17(10), 939-954 (1995)
[17] Guerrout, E., Ait-Aoudia, S., Michelucci, D., Mahiou, R.: Hidden markov random fields and direct search methods for medical image segmentation. In: Proceedings of the 5th International Conference on Pattern Recognition Applications and Methods. pp. 154-161 (2016)