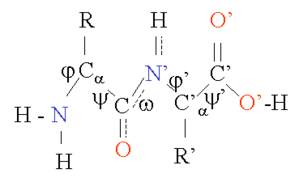
(a)
(b)
(c)
Pictures (b) & (c)taken from [1], Picture (a) taken from
Wiki
|
Preliminaries:
- The protein structure is reduced to a c-alpha trace
i.e. from (a) to (b)
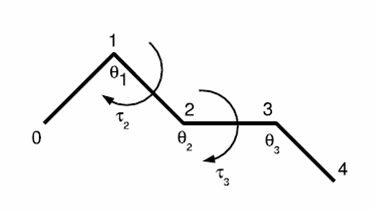
- Now we only have the bond angles (theta) and dihedral
angles (tau) to consider. See (b)
- The bond lengths between two consecutive c-alpha atoms
remain fixed at 3.8 angstroms
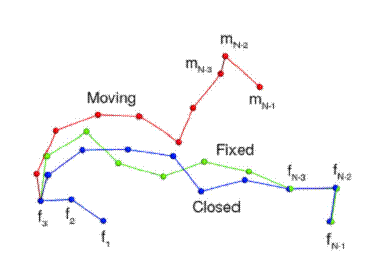
FCCD
- Seeks to move the c-alpha atoms m(N-3), m(N-2) and m(N-1)
in "moving" so as to align them with the c-alpha atoms of
f(N-3), f(N-2) and f(N-1) in "fixed"
- The positions of the first three c-alpha atoms
"moving" is the same as that of the first three c-alpha
in "fixed"
- In order to move the last three atoms of "moving"
onto the last three atoms of "fixed", FCCD iteratively
picks a bond (or vector) along "moving" and calculates
the optimal rotation matrix that will
--rotate all other vectors downstream of it in
the desired direction
--after that reduce the interatomic distance between
the last three atoms of both "moving" and "fixed"
- For a detailed mathematical treatment of FCCD please
consult [1]
|