Please submit both code and writeup online by 12:01am PST on Saturdays, June 4, 2016. Please provide all code (sufficiently commented) so that we can run it ourselves. Submit your writeup as a PDF and limit to 4 pages with reasonable fonts and margins. Please do not forget to write your name on the first page of your writeup.
In this homework you will use the diabetes dataset for running your experiments in problem 1 and 3. For your ease of use, we have provided the links to the libsvm format and the C4.5 format of the same data in problem 1 and problem 3.
1.0 We will be using your ID3 learner (normal and bagged) on the diabetes dataset diabetes dataset. If you suspect your implementation was faulty, you are free to use an existing implementation of these algorithms (e.g. the J48 learner in the Weka Java libraries). Do not use pruning with this problem.
1.1 Modify your ID3 to accept a parameter of maximum depth, beyond which no further splitting is allowed. Consider the ID3 learner alone, speculate whether its bias increase or decrease with this parameter? What about its variance? Why? Verify your conclusion by actually measuring bias and variance for different numbers of maximum depth. At the minimum, try maximum depths of 1, 2, and 3.
1.2 How does the accuracy obtained by ID3 with bagging change as the maximum depth varies? Why? Measure bias and variance in each case. What does this say about your hypothesis?
1.3 To learn more about bias-variance decomposition, see Pedro's ICML paper. Pedro also has a reference implementation. In this homework, generate at least 10 bootstrap samples (30 is recommended if you could afford it) from the original training set and use them when you compute the bias and variance.
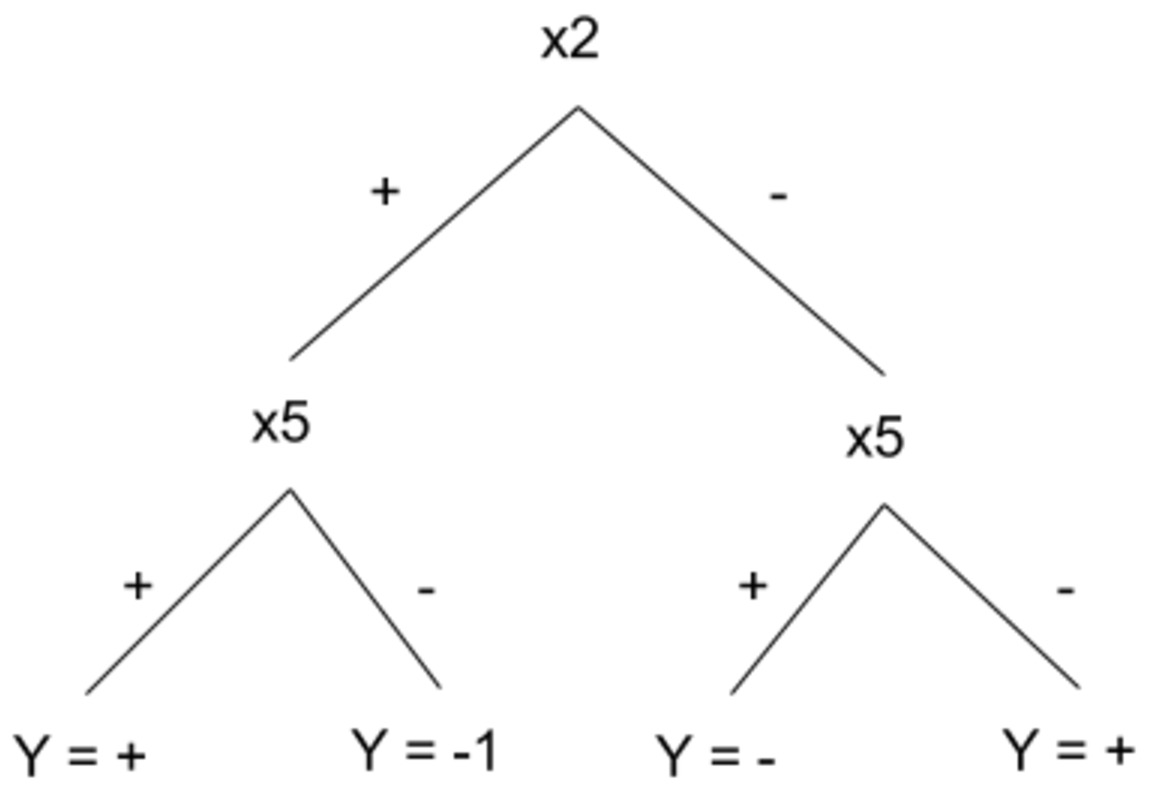
2.0 Suppose we have data with 10-dimensional boolean features [X1,...,X10] which belong to either of Y= + or Y = - classes. We would like to describe our data by a depth-2 regular decision tree. How many examples do we need for training to guarantee with 99% confidence that the learned binary tree has true error of at most 3%?
Appendix: A regular depth-2 decision tree is a decision tree with depth 2 and four leaves and it is required that the left and right child of the root to test the same attribute. An example of a regualr depth-2 decsion tree is shown below. Please note that only two attributes are used for building such a tree.

3.0 What is the VC Dimension of the class of k intervals on the real line. (Hint: Start by considering only one interval and then the union of 2 intervals and justify the generalized case of k intervals. You don’t have to give a formal proof but you must present the key ideas from which the reader could construct a formal proof.)
4.0 Download LIBSVM, currently the most widely used SVM implementation. Peruse the documentations to understand how to use it.
4.1 Download the diabetes dataset.
4.2 Run LIBSVM to classify diabetes. Try different kernels (0-3) and use default parameters for everything else. How does it vary with different choice of kernel?
4.3 Modify your naive bayes classifier from homework 3 to apply to this dataset. How does the accuracy compare to what can be obtained using LIBSVM? Why?
4.4 How do your accuracies compare with that of the bagged decision tree from homework 3? Also, measure the bias and variance of SVMs on the diabetes dataset and compare with bagging's. Explain the difference (or lack of difference).