Due: at 11pm on July 5. Submit via Catalyst CollectIt (a.k.a. Dropbox).
You will be working with real measurements of physical and biological variables from the Puget Sound. These datasets were collected at different times, and are therefore stored in different files. A common task in this kind of context is to integrate several files into one file, then perform some analysis of the result. This integration step is ubiquituous: It is rare that a file organization appropriate for capturing the data is also appropriate for analyzing the data. Some kind of restructuring step is frequently required.
The scientific question we are exploring is related to the NSF award titled Significance of nitrification in shaping planktonic biodiversity in the ocean.
Specifically, we consider the following question: "Which environmental variables correlate with the abundance of Ammonia-Oxidizing Archaea (AOA)?"
From the proposal:
Microorganisms sustain the biogeochemical cycling of nitrogen, one of the most important nutrient cycles on earth. A key step in this cycle, the oxidation of ammonia to nitrite by autotrophic microorganisms, was for a century thought mediated by a few restricted bacterial genera. Significant ammonia oxidation, perhaps most, is now attributed to a previously enigmatic group of Archaea - the ammonia-oxidizing archaea (AOA) - of high abundance in both marine and terrestrial environments. The investigators prior physiological and environmental analyses, the foundation for this proposal, have shown that AOA are active within the marine photic zone and that their competitive fitness in the marine environment is at least in part attributable to an extremely high affinity for ammonia - growing at near maximum growth rates at concentrations of ammonia that would not sustain known bacterial ammonia oxidizers and an unusual copper-based respiratory system that may render them more competitive in iron limited environments. The compelling inference from these prior analyses is that AOA alter and possibly control the forms of fixed nitrogen available to other microbial assemblages within the photic zone by converting ammonia, a nearly universally available form of nitrogen, into nitrite, a form only available to nitrite oxidizing bacteria and some phytoplankton. If correct, this has a significant impact on biodiversity.
An important step in testing this hypothesis is to determine the conditions in which AOA is reponsible for ammonia oxidation; i.e., which environmental variables are associated with high abundance of AOA.
Obtain the files you need by downloading the homework3.zip file and unzipping it to create a homework3 directory/folder. You will do you work by modifying two files — correlate.py and answers.txt — and then submitting the modified versions. Add your name to the top of each of these files.
You will submit answers.txt as a text file. You can edit text files using IDLE. If you choose to edit it using a word processor such as Microsoft Word, be sure to save it as text.
What to turn in: Nothing for Problem 1.
Run the program correlate.py
This program fails with the following error:
File "correlate.py", line 38, in <module>
print pearsons(env_variables, amoas)
File "correlate.py", line 12, in pearsons
xbar = mean(xs)
NameError: global name 'mean' is not defined
The function "pearson" references two functions "mean" and "sample_std" that have not been implemented.
Implement these functions.
Each function will take a single parameter, a list of numbers.
The mean is the sum of the numbers divided by the length of the list.
The sample standard deviation ("sample_std") is the square root of the sum of squared differences.
Use the formula from Wikipedia:
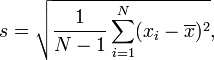
What to turn in: Run your completed program and paste the output of the program into answers.txt
You will convert the code in the section marked PROBLEM 3 into a new
function named extract_pearsons.
This function should take three parameters:
Its return value will be the Pearson coefficient between the data in the two columns.
This function should first open the file named by the filename parameter and read the first line of text containing the column names split it into a list.
Next, the function will need to find which position in the list of column headers contains the value of columnA and
columnB.
You may use the index method for this task,
or you may use a for loop to iterate over the element yourself.
For example,
>>> print ["my", "dog", "is", "nuts"].index("dog")
1
Finally, using the positions of columnA and columnB, you will extract the appropriate columns and compute the Pearson's coefficient.
Once you function is written, add the following lines to the bottom:
print "Correlation of Salinity and Archaeal amoA concentration: "
print extract_pearsons("hood_canal_august_08.csv", "Sal", "arcamoA")
and run the program.
What to turn in: paste the output of the program into answers.txt
Use your new function to find which of the following environmental variables has a
positive correlation with arcamoA in the file hood_canal_august_08.csv.
column_names = [ "NO2 (uM)" ,"Temp (C)" ,"Sal" ,"O2 (mg/L)" ,"Chl (mg/m3)" ,"NO3- (uM)" ,"NH4 (uM)" ,"Sigma-t (Kg/m3)" ,"Transm (%)" ,"PAR" ]
Feel free to copy the list above into your code.
What to turn in: Write the column names that have a positive correlation in answers.txt, one per line. Also turn in the complete program correlate.py
You have been given four files:
Create a new program in a separate file called integrate_hood_canal.py
This program will combine all four input files into one output file called hood_canal.csv
Each input file has the same set of columns. The output should have the same columns as each input file, but include all the data from all four files.
Your result should be indentical to combined.csv
What to turn in: The program integrate_hood_canal.py
Change the program correlate.py to refer to the combined file, and re-run it.
What to turn in: Paste the output of the program correlate.py after changing the file name from hood_canal_august_08.csv to hood_canal.csv.
You are almost done!
At the bottom of your answers.txt file, in the “Collaboration” part, state which students or other people (besides the course staff) helped you with the assignment, or that no one did.
At the bottom of your answers.txt file, in the “Reflection” part, state how many hours you spent on this assignment. Also state what you or the staff could have done to help you with the assignment.
Submit the following files via Catalyst CollectIt (a.k.a. Dropbox):